Maharashtra Board | Class 12 Biology Chapter 4 Molecular Basis of Inheritance Solutions
Class 12 biology textbook Solutions for Class 12, Biology Chapter 4 Molecular Basis of Inheritance Solutions maharashtra state board are provided here with simple step-by-step detailed explanations. These solutions for plant water relation are very popular among Class 12 students for biology chapter 4 Molecular Basis of Inheritance Solutions come handy for quickly completing your homework and preparing for exams.
All questions and answers from the biology textbook Solutions Book of Class 12 biology Chapter 4 are provided here for you for free. You will also love the experience on ybstudy class 12 Solutions. All biology textbook Solutions. Solutions for class 12, These biology textbook solutions are prepared by biology experts and are 100% accurate.
Maharashtra Board | Class 12 Biology Chapter 4 Molecular Basis of Inheritance Solutions
Q. 1 Multiple Choice Questions
1. Griffith worked on ………….
a. Bacteriophage
b. Drosophila
c. Frog eggs
c. Streptococci
2. The molecular knives of DNA are …………..
a. Ligases
b. Polymerases
c. Endonucleases
d. Transcriptase
3. Translation occurs in the ……………
a. Nucleus
b. Cytoplasm
c. Nucleolus
d. Lysosomes
4. The enzyme required for transcription is ………………
a. DNA polymerase
b. RNA polymerase
c. Restriction enzyme
d. RNAase
5. Transcription is the transfer of genetic information from …………..
a. DNA to RNA
b. tRNA to mRNA
c. DNA to mRNA
d. mRNA to tRNA
6. Which of the following is NOT part of protein synthesis?
a. Replication
b. Translation
c. Transcription
d. All of these
7. In the RNA molecule, which nitrogen base is found in place of thymine?
a. Guanine
b. Cytosine
c. Thymine
d. Uracil
8. How many codons are needed to specify three amino acid?
a. 3
b. 6
c. 9
d. 12
9. Which out of the following is NOT an example of inducible operon?
a. Lactose operon
b. Histidine operon
c. Arabinose operon
d. Tryptophan operon
10. Place the following event of translation in the correct sequence
i. Binding of met-tRNA to the start codon.
ii. Covalent bonding between two amino acids.
iii. Binding of second tRNA.
iv. Joining of small and large ribosome subunits.
A. iii, iv, i, ii
B. i, iv, iii, ii
C. iv, iii, ii, i
D. ii, iii, iv, I
Q. 2 Very Short Answer Questions
1. What is the function of an RNA primer during protein synthesis?
Answer : RNA primer acts as a starting point for replication. It is a short segment of RNA complementary to the original strand of DNA. With the help of RNA primer, DNA polymerases can add new nucleotides to an existing strand of DNA.
2. Why the genetic code is considered as commaless?
Answer: Genetic code considered as commaless because it is continuous and non overlapping. Explanation: Genetic code is continuous, commaless and non overlapping, Genetic code contain many codons that are arranged in a particular array to form open reading frame. The genetic code is read as a continuous base sequence.
3. What is genome?
Answer: Total amount of DNA is present in an organism is called as genome.
4. Which enzyme does remove supercoils from replicating DNA?
Answer: Topoisomerase I is a ubiquitous enzyme whose function in vivo is to relieve the torsional strain in DNA, specifically to remove positive supercoils generated in front of the replication fork and to relieve negative supercoils occurring downstream of RNA polymerase during transcription.
5. Why are Okazaki fragments formed on lagging strand only?
Answer: Okazaki fragments form because the lagging strand that is being formed have to be formed in segments of 100–200 nucleotides. This is done DNA polymerase making small RNA primers along the lagging strand which are produced much more slowly than the process of DNA synthesis on the leading strand.
6. When does DNA replication take place?
Answer: DNA replication occurs in the cytoplasm of prokaryotes and in the nucleus of eukaryotes. Regardless of where DNA replication occurs, the basic process is the same.
7. Define term- codon and codogen.
Answer :
- Codon: Triplets of Nucleotide present on m-RNA is called as codon.
- Codogen: Triplets of nucleotide present on DNA is called as codogen.
8. What is degeneracy of genetic code?
Answer: Degeneracy of codons is the redundancy of the genetic code, exhibited as the multiplicity of three-base pair codon combinations that specify an amino acid
9. Which are the nucleosomal ‘core’ histones?
Answer: The nucleosome core particle represents the first level of chromatin organization and is composed of two copies of each of histones H2A, H2B, H3 and H4, assembled in an octameric core with 146-147 bp of DNA tightly wrapped around it.
Q. 3 Short Answer Questions:
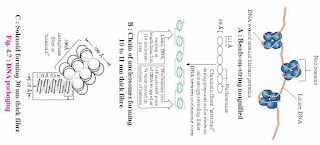
1. Write short note on DNA packaging in eukaryotic cell.
Answer: Length of DNA double helix molecule, in a typical mammalian cell is approximately 2.2 meters. (This can be worked out simply by multiplying the total number of base pairs with distance between the consecutive base pairs).
Packaging in Eukaryotes:
Eukaryotes show well organized nucleus containing nuclear membrane, nucleolus and thread-like material in the form of chromosomes. In the chromosomes, DNA is associated with histone and non-histone proteins as was reported by R. Kornberg in 1974. Histones are the proteins that are rich in lysine and arginine residues.
Both these amino acid residues are basic amino acids and carry positive charges with them (histones + protamine). These histones organize themselves to make a unit of 8 molecules known as histone octamer. The negatively charged helical DNA is wrapped around the positively charged histone octamer, forming a structure known as nucleosome. The nucleosome core is made up of two molecules of each of four types of histone proteins viz. H2A, H2B, H3and H4. H1 protein binds the DNA thread where it enters (arrives) and leaves the nucleosome.
One nucleosome approximately contains 200 base pair long DNA helix wound around it (fig. 4.5). About 146 base pair long segment of DNA remains present in each nucleosome. Nucleosomes are the repeating units of chromatin, which are thread-like, stained (coloured) bodies present in nucleus. These look like ‘beads-on-string’, when observed under an electron microscope. DNA helix of 200 bps wraps around the histone octamer by 1¾ turns.
The chromatin is packed to form a solenoid structure of 30 nm diameter (300A0) and further supercoiling tends to form a looped structure called chromatin fiber, which further coils and condense at metaphase stage to form the chromosomes. The packaging of chromatin at higher levels, need additional set of proteins that are called Non-Histone Chromosomal proteins (NHC).
2. Enlist the characteristics of genetic code.
Answer: Genetic code of DNA has certain fundamental characteristics –
i. Genetic code is a triplet code: Sequence of three consecutive bases constitute codon, which specifies one particular amino acid. Base sequence in a codon is always in 5’ 3’ direction. In every living organism genetic code is a triplet code.
ii. Genetic code has distinct polarity : Genetic code shows definite polarity i.e. direction. It, therefore, is always read in 5’ 3’ direction and not in 3’ 5’ direction. Otherwise message will change e.g. 5’AUG 3’.
iii. Genetic code is non-overlapping : Code is non overlapping i.e. each single base is a part of only one codon. Adjacent codons do not overlap. If non-overlapping, then with 6 consequtive bases only two amino acid molecules will be in the chain. Had it been overlapping type, with 6 bases, there would be 4 amino acid molecules in a chain.
iv. Genetic code is commaless : There is no gap or punctuation mark between successive/ consecutive codons.
v. Genetic code has degeneracy : Usually single amino acid is encoded by single codon. However, some amino acids are encoded by more than one codons. e.g. Cysteine has two codons, while isoleucin has three codons.
vi. Genetic code is universal : By and large in all living organisms the specific codon specifies same amino acid.
vii. Genetic code is non-ambiguous : Specific amino acid is encoded by a particular codon. Alternatively, two different amino acids will never be encoded by the same codon.
viii. Initiation codon and termination codon: AUG is always an initiation codon in any and every mRNA. AUG codes for amino acid methionine. Out of 64 codons, three codons viz. UAA, UAG and UGA are termination codons which terminate/ stop the process of elongation of polypeptide chain, as they do not code for any amino acid.
ix. Universal : Usually in all organisms the specific codon specifies same amino acid.
x. Codon and anticodon : Codon is a part of DNA e.g. AUG is codon. It is always represented as 5’ AUG 3’. Anticodon is a part of tRNA. It is always represented as 3’UAC 5’.
3. Write a note on applications of DNA finger printing.
Answer: every individual has its unique genetic make-up, which may be called its Fingerprint. The technique developed to identify a person with the help of DNA restriction analysis, is known as DNA profiling or DNA fingerprinting. The technique of finger printing was first given by British geneticist, Dr. Alec Jeffreys in 1984. DNA fingerprinting technique is based on identification of nucleotide sequence present in this wonder molecule. About 99.9% of nucleotide sequence in all persons, is same. Only some short sequences of nucleotides differ from person to person.
Application of DNA fingerprinting
1. In forensic science, DNA finger printing is used to solve problems of rape and some complicated murder cases.
2. DNA finger printing is used to find out the biological father or mother or both, of the child, in case of disputed parentage.
3. DNA finger printing is used in pedigree analysis in cats, dogs, horses and humans.
4. Explain the role of lactose in ‘Lac Operon’.
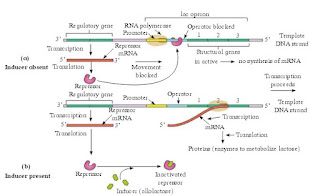
Answer: When lactose is present, the lac genes are expressed because allolactose binds to the Lac repressor protein and keeps it from binding to the lac operator. Allolactose is an isomer of lactose. Small amounts of allolactose are formed when lactose enters E. coli.
Q. 4 Short Answer Questions:
1. Write a note on Human genome project (HGP).
Answer: The human genome project was initiated in 1990 under the International administration of the Human Genome Organization (HUGO). This project was co-ordinated by the US department of Energy and National institute of health. Human Genome Project formally began in 1990 and was completed in 2003.The human genome project is a multinational research project to determine the genomic structure of humans.
The main aims of project are –
I. Mapping the entire human genome at the level of nucleotide sequences.
II. To store the information collected from the project in databases.
III. To develop tools and techniques for analysis of the data.
IV. Transfer of the related technologies to the private sectors, such as industries.
V. Taking care of the legal, ethical and social issues which may arise from project.
2. Describe the structure of ‘Operon’.
Answer : Lac operon : Lactose or lac operon of E.coli is inducible operon.Theoperonisswitchedonwhenachemical inducer- lactose is present in the medium.
Lac operon consists of following components :
1. Regulator gene (repressor gene)
2. Promoter gene
3. Operator gene
4. Structural genes
 |
| Fig : Lac Operon |
1. Regulator gene: This gene controls the operator gene in cooperation with an inducer present in the cytoplasm. Regulator gene preceeds the promoter gene. Regulator gene produces a protein called repressor protein. Repressor binds with operator gene and represses (stops) its action. It is called regulator protein.
2. Promoter gene: This gene preceeds the operator gene. It is present adjacent to operator gene. The promoter gene marks the site at which the RNA Polymerase enzyme binds.
3. Operator gene: It preceeds the structural genes. This controls the functioning of structural genes. It lies adjacent to the Structural genes.
4. Structural gene: There are 3 structural genes in the sequence lac-Z, lac-Y and lac-A. Enzymes produced are E-galactosidase, E-galactoside permease and transacetylase respectively.
Q. 5 Long Answer Questions
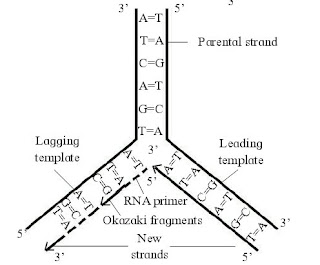
1. Explain the process of DNA replication.
Answer: DNA Replication: The DNA molecule regulates and controls all the activities of the cell. Because of its unique structure, it is able to control the synthesis of other molecules of the cell. At the same time when the cell reproduces, the DNA also should duplicate itself. In eukaryotic organisms, replication of DNA takes place only once in the cell cycle. It occurs in the S- phase of interphase in the cell cycle. DNA replicates through Semiconservative mode of replication.
 |
| Fig : DNA replication. |
1. Activation of Nucleotides:
The four types of nucleotides of DNA i.e. dAMP, dGMP, dCMP and dTMP are present in the nucleoplasm. They are activated by ATP in presence of an enzyme phosphorylase. This results in the formation of deoxyribonucleotide triphosphates i.e. dATP, dGTP, dCTP and dTTP. The process is known as Phosphorylation.
2. Point of Origin or Initiation point:
It begins at specific point ‘O’ -origin and terminates at point ‘T’. Origin is flanked by ‘T’ sites. The unit of DNA in which replication occurs, is called replicon. In prokaryotes, there is noly one replicon.
3. Unwinding of DNA molecule:
Now enzyme DNA helicase operates by breaking weak hydrogen bonds in the vicinity of ‘O. The strands of DNA separate and unwind. This unwinding is bidirectional and continues as ‘Y’ shaped replication fork. Each separated strand acts as template. The two separated strands are prevented from recoiling (rejoining) by SSBP (Single strand binding proteins )
4. Replicating fork:
The point formed due to unwinding and separation of two strands appear like a Y-shaped fork, called replicating/replication fork.
5. Synthesis of new strands:
Each separated strand acts as mould or template for the synthesis of new complementary strand. It begins with the help of a small RNA molecule, called RNA primer. RNA primer get associated with the 3’ end of template strand and attracts complementary nucleotides from surrounding nucleoplasm.
These nucleotides molecules bind to the complementary nucleotides on the template strand by forming hydrogen bonds (i.e. A=T or T=A; G = C or C = G). The newly bound nucleotides get interconnected by phosphodiester bonds, forming a polynucleotide strand. The synthesis of new complementary strand is catalyzed by enzyme DNA polymerase. The new complementary strand is always formed in 5’- 3’ direction.
6. Leading and Lagging strand:
The template strand with free 3’end is called leading template and with free 5’ end is called lagging template. The process of replication always starts at C-3 end of template strand and proceeds towards C-5 end. As both the strands of the parental DNA are antiparallel, new strands are always formed in 5’ to 3’ direction.
One of the newly synthesized strand develops continously towards replicating fork is called leading strand. Another new strand develop discontinuously away from the replicating fork is called lagging strand. DNA synthesis on lagging template takes place in the form of small fragments, called Okazaki fragments.
Okazaki fragments are joined by enzyme DNA ligase. RNA primers are removed by DNA polymerase and replaced by DNA sequence with the help of DNA polymerase-I in prokaryotes and DNA polymerase-I in eukaryotes. Finally, DNA gyrase (topoisomerase) enzyme forms double helix to form daughter DNA molecules.
7. Formation of daughter DNA molecules:
At the end of the replication, two daughter DNA molecules are formed. In each daughter DNA, one strand is parental and the other one is totally newly synthesized. Thus, 50% is contributed by mother DNA. Hence, it is described as semiconservative replication.
2. Describe the process of transcription in protein synthesis.
Answer: The protein synthesis occurs in two steps, namely transcription and translation. During transcription, the information from DNA is encoded into mRNA. During translation, the mRNA works with a ribosome and tRNA to synthesize proteins.
Transcription takes place in the nucleus. During transcription, DNA partially unwinds by the enzyme helicase. This results in the single nucleotide chain to be copied. RNA polymerase reads the DNA strand from the 3′ to 5′ direction and synthesizes the complementary strand of messenger RNA in the 5′ to 3′ direction.
The DNA strand which is transcribed is called as the template strand. The nucleotides on the RNA strand are complementary to the nucleotides on the DNA strand. DNA cytosine pairs with RNA guanine, DNA guanine pairs with RNA cytosine, DNA thymine pairs with RNA adenine and DNA adenine pairs with RNA uracil. After mRNA is synthesized, it is transported to the cytoplasm where it binds with ribosomes.
3. Describe the process of translation in protein synthesis.
Answer:
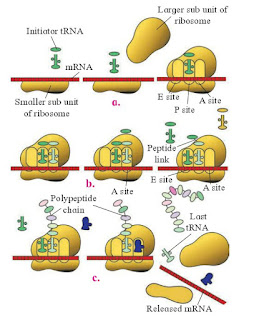 |
| Fig : Translation a-Initiation, b-Elongation, C-Termination |
Ribosomes are the sites of protein synthesis. Ribosomes have three important binding sites, one for mRNA and two (A site and P site) for tRNA. The start codon methionine occupies the P site and the second codon occupies the A site.
The tRNA molecule whose anticodon is complementary to the mRNA forms a base pair with the mRNA in the A site. A peptide bond is formed between the amino acid attached to the tRNA in the A site and the methionine in the P site. The ribosome then slides down the mRNA in such a way that the tRNA in the A site moves to the P site and a new codon occupies the A site.
This process continues until one of the three stop codons occupies the A site. At that point, the protein chain connected to the tRNA in the P site is released and the translation is complete.
4. Justify the statements. If the answer is false, change the underlined word(s) to make the statement true.
i. The DNA molecule is double stranded and the RNA molecule is single stranded.
Answer: True
ii. The process of translation occurs at the ribosome.
Answer: True
iii. The job of mRNA is to pick up amino acids and transport them to the ribosomes.
Answer: True
iv. Transcription must occur before translation may occur.
Answer: True
Chapterwise Textbook Solutions:
| Chapter 2: Reproduction in Lower & Higher Animals |
| Chapter 3: Inheritance and Variation |
| Chapter 4: Molecular Basis of Inheritance |
| Chapter 6: Plant Water Relation |
| Chapter 7: Plant Growth and Mineral Nutrition |
| Chapter 8: Respiration and Circulation |
| Chapter 9: Control and Coordination |
| Chapter 10: Human Health and Diseases |
| Chapter 11: Enhancement of Food Production |
| Chapter 12: Biotechnology |
| Chapter 13: Organisms and Populations |
| Chapter 14: Ecosystems and Energy Flow |